ESR10: Development of new drugs to inhibit the neurodegenerative process in dopamine neurons
Aoife Bannon (University of Siena)
Supervisor: Massimo Valoti
Project summary
The development of biomarkers for the diagnosis and monitoring disease progression in Parkinson's disease (PD) is of great importance and may prove to be useful in early diagnosis and in identifying at-risk individuals and subgroups of PD. Recently, there has been increased interest in the alteration of lipid metabolism in PD and linking dysregulation of lipid homeostasis to the disease. Circulating lipids are involved in many complex disease processes, and plasma lipid profiling has been used to identify lipidomic biomarkers associated with various diseases. The general aim of my research project is to employ these kind of studies to PD i.e. generate a lipidomic map of the disease. In order to achieve this goal, I will first carrying out in-vitro experiments and investigating lipid alterations in cell models of PD. Cellular models will be treated with PD inducers or neurotoxins will be compared to cells treated with neuroprotective agents. Using lipid extraction and mass spectrometry procedures I hope to discover changes in lipid metabolism and identify lipid species that are associated with the disease state. If this part of the project is successful, these lipid species will be investigated as plasma biomarkers of PD in vivo and their interactions with newly developed drugs may be investigated.
The specific aims of the project are:
- Optimise lipid extraction procedures on SHSY-5Y neuroblastoma cells for mass spectrometric analysis
- Identify alterations in lipid species present in PD cell models
- Investigate plasma lipid alterations in vivo
- Investigate the lipid-drug interactions
Results [top]
The lipid profile of undifferentiated SHSY5Y neuroblastoma cells was obtained using a newly developed lipid extraction method (MMC method) and Q-TOF LC/MS analysis.
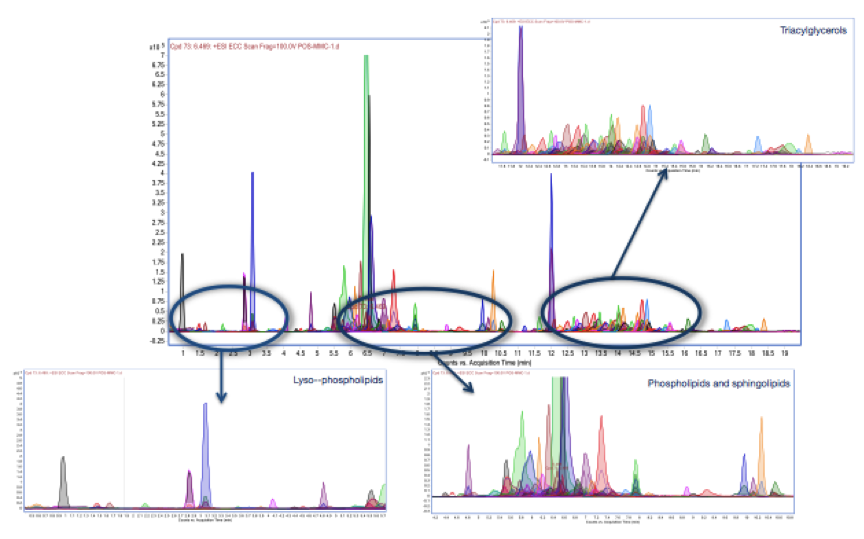
Figure 1. Lipid Profile of undifferentiated SH-SY5Y cells following MMC extraction and Q-TOF LC/MS analysis.
Preliminary trials suggest that the new lipid extraction method provides satisfactory recovery of lipids of cell samples compared to the gold standard methods (Folch and Bligh & Dyer). This should provide a basic profile against which cells treated with toxic/protective agents can be compared.
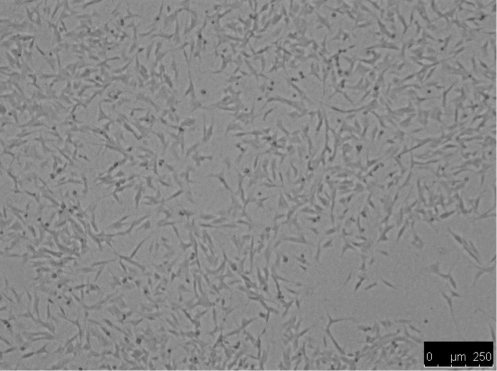
Differentiation was induced by treating SHSY5Y cells with 10uM RA in the dark for 7 days resulting in morphological differences from undifferentiated cells, such as elongation and neurite outgrowth.
 |
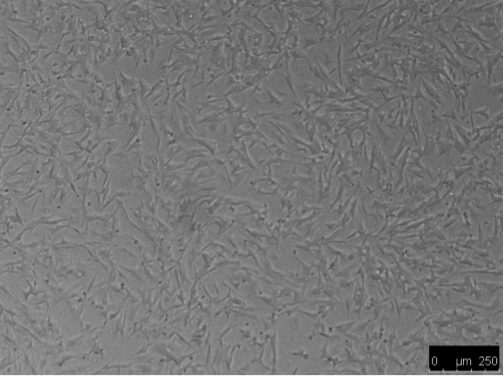 |
Figure 2. SHSY5Y cells: undifferentiated (left) vs differentiated (right)
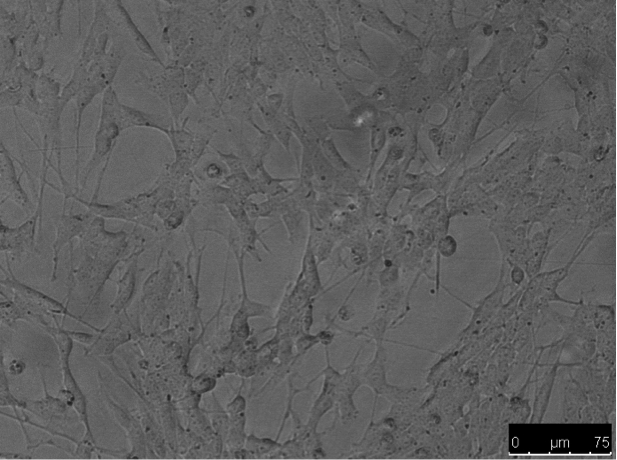
Figure 3. Differentiated SHSY5Y cells.
Outputs [top]
I am currently assisting in the writing of a review entitled "Interactions of Monoamine Oxidase Inhibitors with Cytochrome P450 enzymes"
which is anticipated to be published within the next few months.
Training [top]
(i) Local level
I have received training in the following laboratory techniques and protocols;
- Cell Culture; general technique and specific protocol for SHSY-5Y cells
- Cell Viabilty Assays; MTT assays
- Fluorescence Microscopy; using acridine orange dye
- Lipid Extraction; Folch method and MMC method
- Mass Spectrometry
- RNA Isolation
- Functional Proteomics
- From Bench to Bedside: Principles of Vaccines R & D
- Drug Design for Monoamine Oxidase Inhibitors: Enzymology, Mitochodria, Neuropathology
(ii) Network level
I have attended all the TINTIN training courses. They are listed below.
- NMR Bootcamp
This course was a basic introduction to nuclear magnetic resonance (NMR) spectroscopy. The lectures included a brief introduction to the fundamental concepts and quantum mechanics behind the technique, an explanation of the different types of NMR spectroscopy, as well as examples of its uses and applications. More in-depth introductions to the applications of NMR in various fields were also given, including peptide NMR, nucleic acid NMR, the use of NMR in metabolomics and in relaxometry and MRI. There were practical classes which demonstrated NMR analysis of small molecules, solving the resulting data, and the use of CRAFT software in metabolomics and analaysis. - Fluorescence and Electron Microscopy Imaging of Cells
This course covered topics in advanced light microscopy, including the use and application of electron microscopy for biological imaging. It involved introductory lectures covering microscopic principles, as well as demonstrations and training in sample preparation, processing, viewing and analysis. - Neuronal Cell Metabolism
This course involved an introductory lecture explaining the alterations in fundamental metabolism in cancer and other diseases, followed by a practical class demonstrating the use of Seahorse analysis to evaluate changes in metaboliam. - Training in Mitochondrial and Cellular Respiratory Physiology
This course was given concurrently with the Neuronal Cell Metabolism course, and involved training on the use of the Oroborus instrument (Oxygraph-2k respirometer) in measuring oxygen consumption rates and hydrogen peroxide production.
Outreach [top]
I gave a presentation on my research during my secondment at Molecular Discovery in Perugia, during which I explained the TINTIN training network and described my part as an ESR in the programme.