ESR9:
In silico reconstruction of the dopamine transporter and design of new drugs with neuroprotective properties
Teodora Djikic (Kadir Has University)
Supervisor: Kemal Yelekci
Project summary
Dopamine transporter (DAT) plays an important role in the termination of neurotransmission by rapid reuptake of dopamine (DA) from the synaptic cleft into presynaptic terminals where it is degraded by monoamine oxidase (MAO). With the blockade of DAT, level of dopamine in synaptic cleft, as well as dopaminergic neurotransmission, will increase, followed by decreases in intracellular degradation and production of reactive oxygen species (ROS) and oxidative stress.
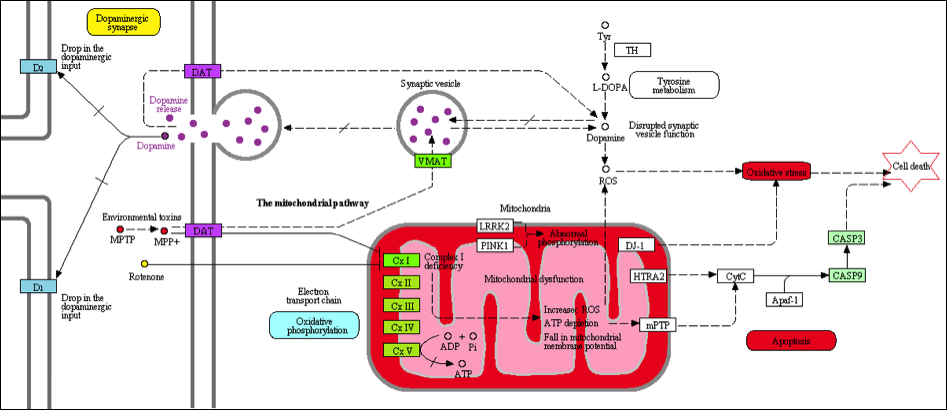
[Adapted from: http://www.kegg.jp/kegg-bin/show_pathway?hsa05012]
The specific aims of the project are:
- Modelling of human dopamine transporter (hDAT)
- Understanding the difference between substrate and inhibitor binding
- Understanding the mechanisms of mitochondrial dysfunction and neuronal cell death
- Designing hDAT substrates or inhibitors with neuroprotective properties
- Synthesis and testing
Results [top]
The first eukaryotic X-ray structure of DAT from Drosophila (dDAT) has been published recently (Penmatsa, 2013).
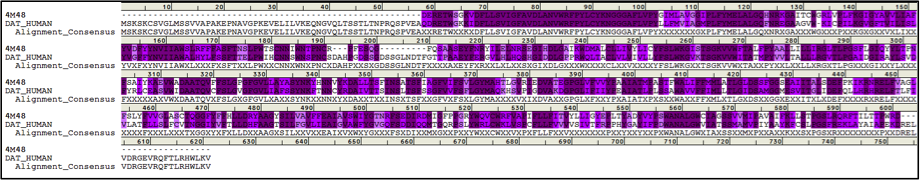
Sequence identity between these two proteins is 49.6% and sequence similarity is 69.2%.
We have created a new model of hDAT, based on dDAT, and minimized it in the environment of the lipid bilayer and solvent water. Molecular dynamics (MD) simulations on an empty transporter revealed small conformational changes of the backbone, but important changes of the amino acid sidechains, especially Asp79, Phe326 and Phe320, thus creating a partially closed state. The transporter was validated with known inhibitors downloaded from CHEMbl database, in Auto Dock 4.2. and GOLD 5 docking software both with rigid protein and with previously mentioned amino acids set to flexible. The obtained results correlated positively with the existing experimental results. The binding free energy for DA, according to pre-existent experiments, is ΔGexperimental = -7.4 kcal/mol; while ΔGcalculated = -6.4 kcal/mol was obtained through our studies. The binding position of DA, also, coincides with previous literatures: interaction with Phe320, Asp79 and Thr156 amino acids. Energies of bindings of inhibitors were smaller than ΔGdopamine (for cocaine: Ki(experimental) = 690 nM, ΔGcalculated = -7.47 kcal/mol, Ki(calculated)= 3.32 μM; and for nortriptyline ΔGcalculated - 8.49 kcal/mol, KKi(calculated) = 596 nM Ki(experimental) = 156 nM) which was expected. Also inhibitors are showing different bindings for the transporter, resulting in interactions with Phe326, Phe320, Ile484, Tyr156, Val152, Asp476 and Ala81.
Molecular dynamic simulations with other substrates are currently running, in order to help us understand the conformational changes of the transporter, as well as mechanisms of its binding and release.
The classic alternating access model implies that the transporter protein takes at least three conformational states during the transport phase:
- Outward-open conformation (the substrate binding pocket is accessible to the extracellular medium)
- Occluded conformation (access to the pocket is blocked from both sides)
- Inward-facing (the pocket is open to the intracellular medium) (Kristensen, 2011).
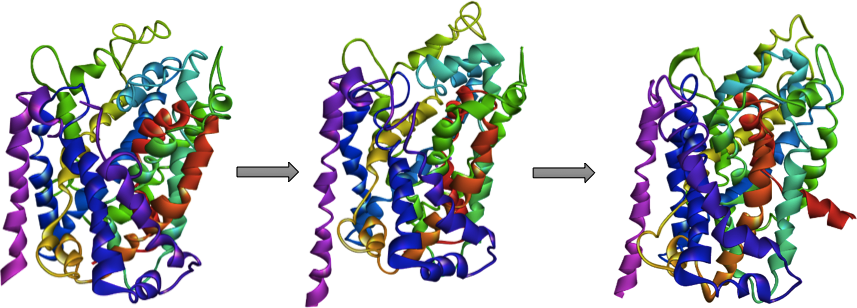
| We hypothesised that inhibitors are binding in the outward-open conformation and blocking the transporter, while substrates are binding to both, but having better scores when binding for the occluded state. Knowing this we would be able to determine whether a compound will be an inhibitor or a substrate, with only docking calculations, and we would be able to use occluded (substrate-bonded) conformation for structure based drug design of neuroprotective DAT substrates. |
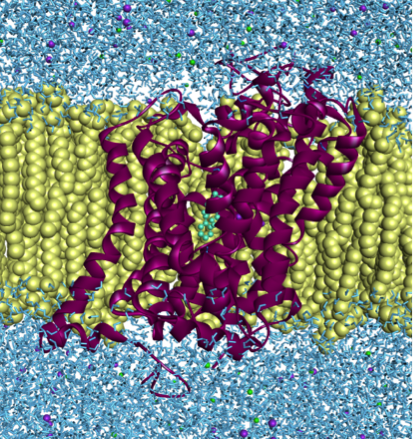
|
Since rasagiline, an irreversible inhibitor of monoamine oxidase-B which can be used as a therapy in Parkinson's disease, has shown some neuroprotective properties, we are also investigating its effects on MAO-A and MAO-B enzymes.
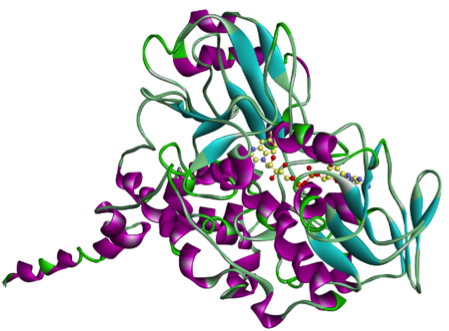
|
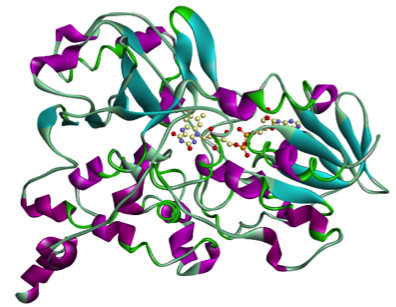
|
| MAO-A | MAO-B |
Outputs [top]
- Peer reviewed publications
- Evranos-Aksö, B., Baysal, İ, Yabanoğlu-Çiftçi, S., Djikic, T.,
Yelekçi, K., Uçar, G. and Ertan, R. (2015)
Synthesis and screening of human monoamine oxidase-A: inhibitor effect of new 2-pyrazoline and hydrazone derivatives.
Arch. Pharm. Chem. Life Sci., 348, 743–756.
[DOI: 10.1002/ardp.201500212]
- Evranos-Aksö, B., Baysal, İ, Yabanoğlu-Çiftçi, S., Djikic, T.,
Yelekçi, K., Uçar, G. and Ertan, R. (2015)
Synthesis and screening of human monoamine oxidase-A: inhibitor effect of new 2-pyrazoline and hydrazone derivatives.
Arch. Pharm. Chem. Life Sci., 348, 743–756.
- Meeting abstracts
- "In silico modelling of human dopamine transporter and design of novel inhibitors" Kemal Yelekci, Teodora Djikic at RICT 2015, International Conference on Medicinal Chemistry
- "Novel reversible and selective pyrazoline based hMAO-A inhibitors: synthesis, docking studies and biological evaluation" B. Evranos-Aksöz, İ. Baysal, S. Yabanoglu Ciftci, T. Djikic, K. Yelekçi, G. Uçar 11th International Symposium on Pharmaceutical Sciences (ISOPS-11) Ankara, Turkey, June 09-12, 2015. "Computer-Aided Drug design for Neurological Targets" Katarina Nikolic, Lazaros Mavridis, Teodora Djikic, Oscar M. Bautista-Aguilera, José Marco-Contelles, Holger Stark, Maria do Carmo Carreiras, Ilaria Rossi, Paola Massarelli, Danica Agbaba, Kemal Yelekci, Rona R. Ramsay, and John B. O. Mitchell
Training [top]
(i) Local level
I have completed all of the obligatory courses for PhD studies successfully. The courses are listed below:
- Advanced bioinformatics
- Object-orientated programing
- Advanced Cell Biology
- Molecular Modeling, Simulation and Visualization
- Computational Systems Biology
- Advanced Computational structural Biology
- Computational Drug Design Techniques
- Computational Neuroscience by University of Washington
- The Brain and Space by Duke University
- Drug Discovery, Development & Commercialization by University of California, San Diego
NMR Mini Boot Camp of BioBank Analyses and Metabolomics Transformation (November 2014)
Fluorescence and electron microscopy imaging of cells (5-8 May 2015)
Outreach [top]